Can I run workflows on an HPC cluster?#
Short answer: yes, you can.
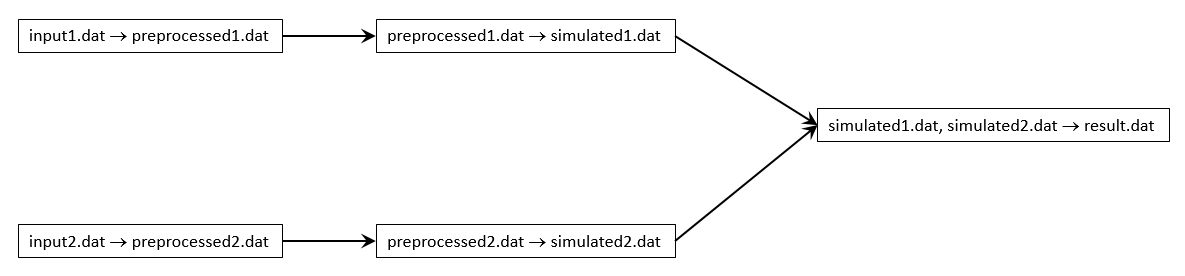
Some workflows do not lend themselves to be completed in a single job. As a running example, consider the following simple workflow:
preprocess first data file
The data in file
input1.dathas to be preprocessed using an applicationpreprocessthat runs only on a single core, and takes 1 hour to complete. The processed data is written to a filepreprocessed1.dat.main computation on first preprocessed file
The preprocessed data in
preprocessed1.datis now used as input to run a large scale simulation using the applicationsimulatethat can run on 20 nodes and requires 3 hours to complete. It will produce an output filesimulated1.dat.preprocess second data file
The data in file
input2.dathas to be preprocessed using an applicationpreprocessthat runs only on a single core, and takes 1 hour to complete. The processed data is written to a filepreprocessed2.dat.main computation on second preprocessed file
The preprocessed data in
preprocessed2.datis now used as input to run a large scale simulation using the applicationsimulatethat can run on 20 nodes and requires 3 hours to complete. It will produce an output filesimulated2.dat.postprocessing data
However, the files
simulated1.datandsimulated2.datare not suitable for analysis, they have to be postprocessed using the applicationpostprocessthat can run on all cores of a single node, and takes 1 hour to complete. It will produceresult.datas final output.
This workflow can be executed using the job script below:
#!/usr/bin/env bash
#PBS -l nodes=20:ppn=36
#PBS -l walltime=10:00:00
cd $PBS_O_WORKDIR
preprocess --input input1.dat --output preprocessed1.dat
simulate --input preprocessed1.dat --output simulated1.dat
preprocess --input input2.dat --output preprocessed2.dat
simulate --input preprocessed2.dat --output simulated2.dat
postprocess --input simulated1.dat simulated2.dat --output result.dat
Just to be on the safe side, 10 hours of walltime were requested rather than the required 9 hours total. We assume that compute nodes have 36 cores.
The problem obviously is that during 1/3th of the execution time, 19 out of the 20 requested nodes are idling. The preprocessing and postprocessing step run on a single node. This wastes 57 node-hours out of a total of 180 node-hours, hence your job has an efficiency of at most 68 %, which is unnecessarily low.
Rather than submit this as a single job, it would be much more efficient to submit it as five separate jobs, two for preprocessing, two for the simulation, the fifth for postprocessing.
Preprocessing the first input file would by done by
preprocessing1.pbs:#!/usr/bin/env bash #PBS -l nodes=1:ppn=1 #PBS -l walltime=1:30:00 cd $PBS_O_WORKDIR preprocess --input input1.dat --output preprocessed1.dat
Preprocessing the second input file would by done by
preprocessing2.pbs, similar topreprocessing1.pbs.The simulation on the first preprocessed file would by done by
simulation1.pbs:#!/usr/bin/env bash #PBS -l nodes=20:ppn=36 #PBS -l walltime=6:00:00 cd $PBS_O_WORKDIR simulate --input preprocessed1.dat --output simulated1.dat
The simulation on the second preprocessed file would by done by
simulation2.pbsthat would be similar tosimulation1.pbs.The postprocessing would by done by
postprocessing.pbs:#!/usr/bin/env bash #PBS -l nodes=1:ppn=36 #PBS -l walltime=6:00:00 cd $PBS_O_WORKDIR postprocess --input simulated1.dat simulated2.dat --output result.dat
However, if we were to submit these three jobs independently, the scheduler may
start them in any order, so the postprocessing job might run first, and immediately
failing because the file simulated1.dat and/or simulated2.dat don’t exist
yet.
Note
You don’t necessarily have to create separate job scripts for similar tasks since it is possible to parameterize job scripts.
Job dependencies#
The scheduler supports specifying job dependencies, e.g.,
a job can only start when two other jobs completed successfully, or
a job can only start when another job did not complete successfully.
Job dependencies can effectively solve our problem since
simulation1.pbsshould only start whenpreprocessing1.pbsfinishes successfully, andsimulation2.pbsshould only start whenpreprocessing2.pbsfinishes successfully, andpostprocessing.pbsshould only start when bothsimulation1.pbsandsimulation2.pbsfinished successfully.
It is easy to enforce this using job dependencies. Consider the following sequence of job submissions:
$ preprocessing1_id=$(qsub preprocessing1.pbs)
$ preprocessing2_id=$(qsub preprocessing2.pbs)
$ simulation1_id=$(qsub -W depend=afterok:$preprocessing1_id simulation1.pbs)
$ simulation2_id=$(qsub -W depend=afterok:$preprocessing2_id simulation2.pbs)
$ qsub -W depend=afterok:$simulation1_id:$simulation2_id postprocessing.pbs
The qsub command returns the job ID, and this is assigned to a bash variable.
It is used in subsequent submissions to specify the job dependencies using
-W depend. In this case, follow-up jobs should only be run when the
previous jobs succeeded, hence the afterok dependencies.
The scheduler can run preprocessing1.pbs and preprocessing2.pbs concurrently
if the resources are available (and can do so on the same node). Once either is done,
it can start the corresponding simulation, again potentially concurrently if 40 nodes
would happen to be free. When both simulations are done, the postprocessing can start.
Since each step requests only the resources it really requires, efficiency is optimal, and the total time could be as low as 5 hours rather than 9 hours if ample resources are available.
Types of dependencies#
The following types of dependencies can be specified:
- afterok
only start the job when the jobs with the specified job IDs all completed successfully.
- afternotok
only start the job when the jobs with the specified job IDs all completed unsuccessfully.
- afterany
only start the job when the jobs with the specified job IDs all completed, regardless of success or failure.
- after
start the job as soon as the jobs the the specified job IDs have all started to run.
A similar set of dependencies is defined for job arrays, e.g.,
afterokarray:<job_id>[] indicates that the submitted job can only start
after all jobs in the job array have completed successfully.
The dependency types listed above are the most useful ones, for a complete list, see the official qsub documentation. Unfortunately, not everything works as advertized.
To conveniently and efficiently execute embarrassingly parallel parts of a workflow (e.g., parameter exploration, or processing many independent inputs), the worker framework or atools will be helpful.
Job success or failure#
The scheduler determines success or failure of a job by its exit status:
if the exit status is 0, the job is successful,
if the exit status is not 0, the job failed.
The exit status of the job is strictly negative when the job failed because, e.g.,
it ran out of walltime and was aborted, or
it used too much memory and was killed.
If the job finishes normally, the exit status is determined by the exit status of the job script. The exit status of the job script is either
the exit status of the last command that was executed, or
an explicit value in a bash
exitstatement.
When you rely on the exit status for your workflow, you have to make sure that the exit status of your job script is correct, i.e., if anything went wrong, it should be strictly positive (between 1 and 127 inclusive).
Note
This illustrates why it is bad practice to have exit 0 as the last
statement in your job script.
In our running example, the exit status of each job would be that of the last
command executed, so that of preprocess, simulate and postprocess
respectively.
Parameterized job scripts#
Consider the two job scripts for preprocessing the data in our running example.
The first one, preprocessing1.pbs is:
#!/usr/bin/env bash
#PBS -l nodes=1:ppn=1
#PBS -l walltime=1:30:00
cd $PBS_O_WORKDIR
preprocess --input input1.dat --output preprocessed1.dat
The second one, preprocessing2.pbs is nearly identical:
#!/usr/bin/env bash
#PBS -l nodes=1:ppn=1
#PBS -l walltime=1:30:00
cd $PBS_O_WORKDIR
preprocess --input input2.dat --output preprocessed2.dat
Since it is possible to pass variables to job scripts when using qsub, we
could create a single job script preprocessing.pbs using two variables
in_file and out_file:
#!/usr/bin/env bash
#PBS -l nodes=1:ppn=1
#PBS -l walltime=1:30:00
cd $PBS_O_WORKDIR
preprocess --input "$in_file" --output "$out_file"
The job submission to preprocess input1.dat and input2.dat would be:
$ qsub -v in_file=input1.dat,out_file=preprocessed1.dat preprocessing.pbs
$ qsub -v in_file=input2.dat,out_file=preprocessed2.dat preprocessing.pbs
Using job dependencies and variables in job scripts allows you to define quite sophisticated workflows, simply relying on the scheduler.